When preparing the jisc collections data for ingestion into OpenAPC in September 2017, more than 8000 articles unfortunately had to be excluded since no DOI had been supplied for them. However, a closer investigation revealed that a significant share of those entries, while lacking persistent identifiers, did include the title of the article (about 3000).
In principle, knowledge of an article title allows for rather easy retrival of the associated DOI: It usually involves copy-pasting the string into a search engine, following the first link to a landing page or full text PDF and looking for the DOI there. While this is doable for a small number of data points, it quickly becomes infeasible for larger sets of data, and 3000 articles is clearly too much to look up manually. Fortunately, there is a solution: At OpenAPC, we usually employ the Crossref REST API to import metadata like ISSNs, journal title or publisher using the DOI as query (that’s why we put so much emphasis on this identifier). However, the API can also be used to search various other metadata fields - like the title of an article.
Let’s have a look at an example from those 3000 articles in question: We want to look up the DOI for an article named
“The phosphorylation of Hsp20 enhances its association with amyloid-<beta> to increase protection against neuronal cell death”
The according query to the Crossref API looks as follows:
This returns a quite comprehensive JSON structure. Using the URL parameter rows=5 will ensure that a maximum of 5 results will be delivered, which is more than sufficient - usually the first one will already be what we are looking for, as it is the case here:
` “DOI”: “10.1016/j.mcn.2014.05.002”, `
` […] `
` “title”: [“The phosphorylation of Hsp20 enhances its association with amyloid-\u03b2 to increase protection against neuronal cell death”] `
However, taking a look at the title string reveals a problem - the spelling in Crossref might differ from our search title (Here the greek letter β is correctly encoded in Unicode in Crossref, but written out as a word in our data). This is an obstacle for automated lookup, because a simple string comparison will fail in these cases. The solution is to use a string metric like the Levenshtein distance which provides a measure of difference between two character sequences. In our case we employ a so-called Levenshtein ratio (LR), which is the Levenshtein distance, normalised to string length. A LR of 1.0 would indicate that the two strings are identical, while a LR 0.0 would mean that they are totally different. In our example, the LR between the Crossref title and our search title is 0.97, which indicates a high level of similarity.
Now we have everything at hand to put together a script for automated reverse DOI lookup which does the following:
For every DOI-less article with a title:
- query the Crossref API, using the article title as search string
- for every returned result, calculate the Levenshtein ratio (LR) between the query title and the result title
- Take the result with the highest LR and do the following:
- If it’s higher than 0.9: Assume identity and import the associated DOI.
- If it’s between 0.8 and 0.9: Present the strings to the user and ask what to do.
- If it’s lower than 0.8: Discard results.
There’s no deeper magic behind those numbers, the 0.8 and 0.9 thresholds simply turn out to work pretty well in practice.
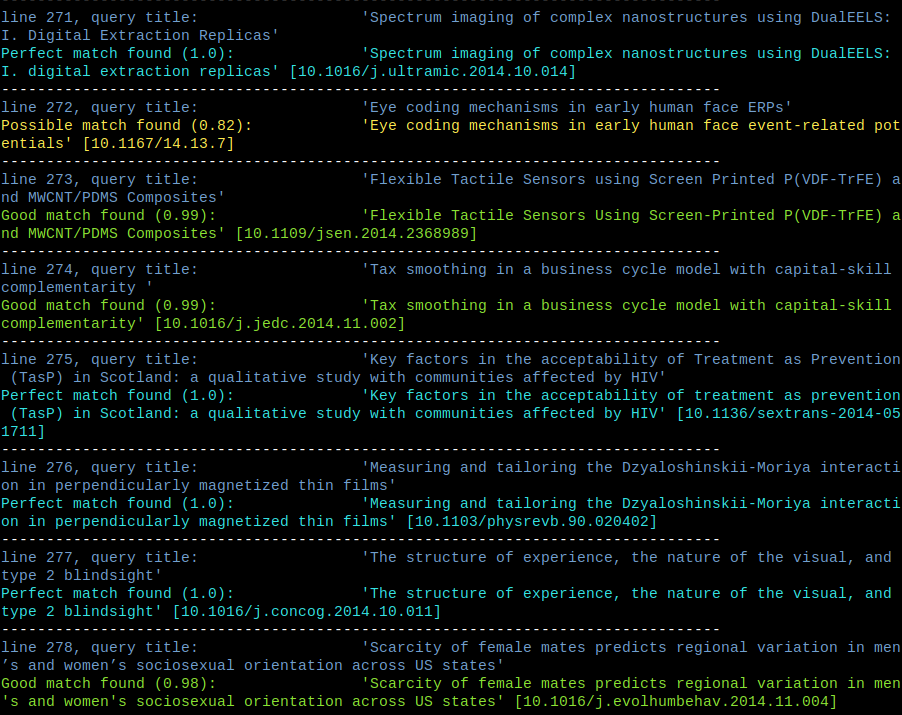
While the whole process took some time, results were very convincing. Here’s a breakdown for the 2970 articles in question:
| match type | Levenshtein ratio | count | percent |
|---|---|---|---|
| perfect | 1.0 | 1469 | 49.5 |
| good | between 1.0 and 0.9 | 935 | 31.5 |
| possible | between 0.9 and 0.8 | 114 | 3.8 |
| no match | lower than 0.8 | 452 | 15.2 |
Altogether, this method could retrieve more than 2400 DOIs automatically. In the end, this led to a net ingestion of 1520 new articles into the OpenAPC core data file.
As always, the python code has been put under an MIT license, so feel free to reuse and experiment!